![]() [Higo group]
[Higo group]
Analysis on the structural polymorphism of IDPs by computer simulation
Group member : Makoto Kikuchi (Professor, Osaka University)
Group member : Mitsunori Takano (Associate Professor, Waseda University)
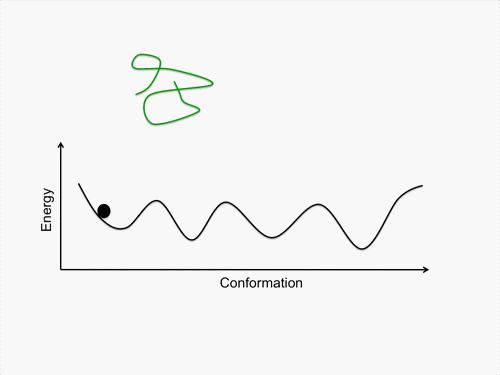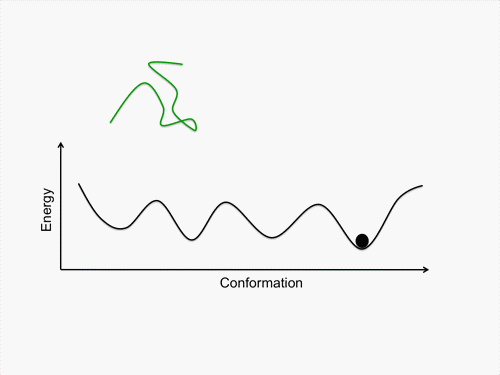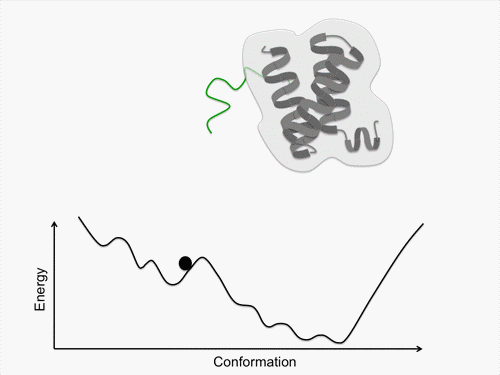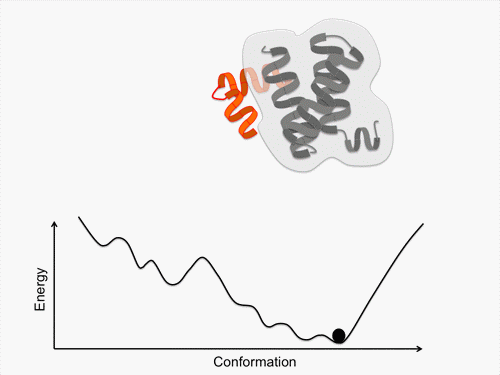
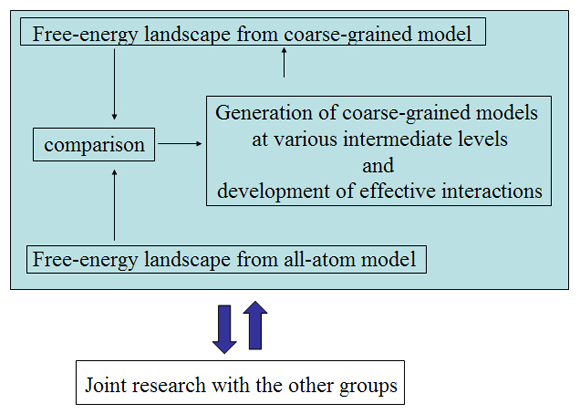
Initially, we perform conformational sampling of the system, which was expressed with an all-atom model in explicit water, to obtain an accurate free-energy landscape. We then use a coarse-grained model to obtain the coarse-grained free-energy landscape. In addition, we develop intermediately grained models to connect the free-energy landscapes from the coarse-grained and the all-atom models. Through this computational process, we are able to predict a picture of the IDP which is consistent with experimental results.
Exhaustive sampling of protein conformation is generally difficult for conventional simulation techniques. We perform a multicanonical method for the conformational sampling: multicanonical molecular dynamics (McMD) simulation for the all-atom model, and multicanonical Monte Carlo (McMC) simulation for the coarse grained models. We produce the free-energy landscape at room temperature through the statistical analysis on the obtained simulation data. Furthermore, we are uncovering the molecular recognition mechanism of IDPs by comparing our results with those from experiment.