Project
![]() [Sato group]
[Sato group]
Development of new methods to determine highly flexible IDP structures
Group leader : Mamoru Sato (Professor, Yokohama City University)
Group member : Toshio Ando (Professor, Kanazawa University)
Group member : Toshio Ando (Professor, Kanazawa University)
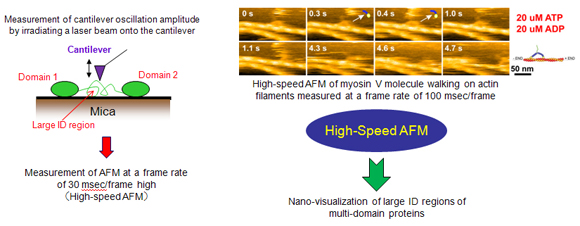
The highly dynamic structures of IDPs, and multi-domain proteins that include large ID regions, cannot be determined by such conventional methods as X-ray crystallography and electron microscopy. Our research group aims to develop further the novel methods which we invented to determine such structures by MD-SAXS and high-speed AFM.
Small-angle X-ray scattering (SAXS) is suitable to determine low-resolution structures of proteins and protein complexes in solution, but the structural data on protein dynamics are averaged over the structural ensemble in protein solution. Therefore we have developed a novel method, named MD-SAXS, which combines SAXS and molecular dynamics (MD) simulation to analyze protein dynamics in solution. This method is especially useful for multi-subunit protein complexes and multi-domain proteins.
Small-angle X-ray scattering (SAXS) is suitable to determine low-resolution structures of proteins and protein complexes in solution, but the structural data on protein dynamics are averaged over the structural ensemble in protein solution. Therefore we have developed a novel method, named MD-SAXS, which combines SAXS and molecular dynamics (MD) simulation to analyze protein dynamics in solution. This method is especially useful for multi-subunit protein complexes and multi-domain proteins.

We have also developed high-speed AFM (atomic force microscopy) and succeeded in visualizing biomolecular processes such as myosin V molecule walking on actin filaments. This method has further been applied to visualize large ID regions of multi-domain proteins, and found to be a suitable method to analyze single molecule dynamics of large ID regions.